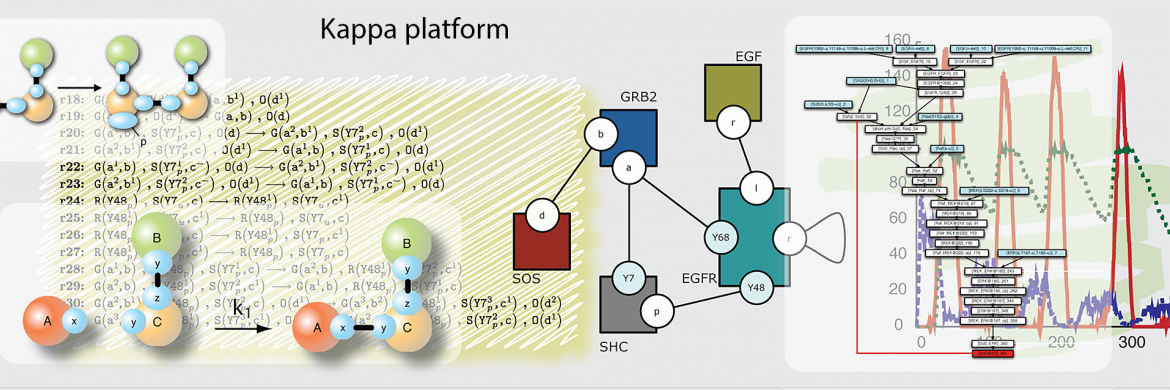
Molecular biology offers breathtaking views of the parts and processes that undergird life and its evolution. It is vexing, then, that we seem unable to analytically grasp the principles that would make the nature of cellular phenotypes more intelligible and their control more deliberate. One can always blame insufficient knowledge, but we also entertain the idea that physics and chemistry need formal and conceptual enrichment from computer science to become an appropriate foundation for systems biology. This view arises from the belief that computation is a natural phenomenon, like gravity or boiling water. We need adequate formalisms and models to reason about computation in the wild.
This view guides many of our lab's interests, which span the development and application of rule-based formalisms for modeling complex systems of molecular interaction, causality in concurrent systems, the interplay between network growth and network dynamics, phenotypic plasticity and evolvability, learning, and aging. Our approach is computational and theoretical. In the past we also conducted experimental work using C. elegans as a model system. Outside collaborations are essential to our group. The size of our team can fluctuate considerably, as we chase grants in pursuit of our passions, not opportunistically. Read more about our research.
The members and collaborators of our Lab typically come from diverse backgrounds, such as molecular biology and genetics, mathematics, computational biology, physics, chemistry, engineering, and computer science. In this diversity we establish what science historian Peter Galison called in his 1997 book Image and Logic a "trading zone", in which we "exchange fish for baskets, enforc[e] subtle equations of correspondence between quantity, quality, and type, and yet utterly disagree on the broader [...] significance of the items exchanged".
Welcome.